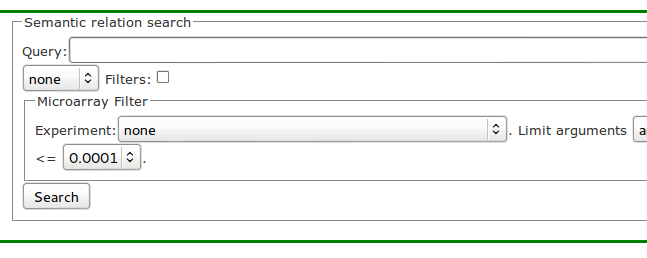
In the interface shown in Figure 1, the user enters the question in the Query field then presses the Search button and gets a list of semantic relations as answers.

Right now, the user question (query) cannot be entered in natural language form. In general, the question is specified as a template (subject, relation, object), which refers to various components of the stored semantic relations. At least one component must be specified, but it is possible to specify two or even all three, depending on the question. The question is forwarded to Lucene which means that full Lucene query syntax is allowed.
A question containing only one of the arguments, e.g. Alzheimer's disease is very general and means any relation between Alzheimer's disease and some other biomedical concepts. Such question will produce as an answer a set of semantic relations that can be used for a quick overview of a concept.
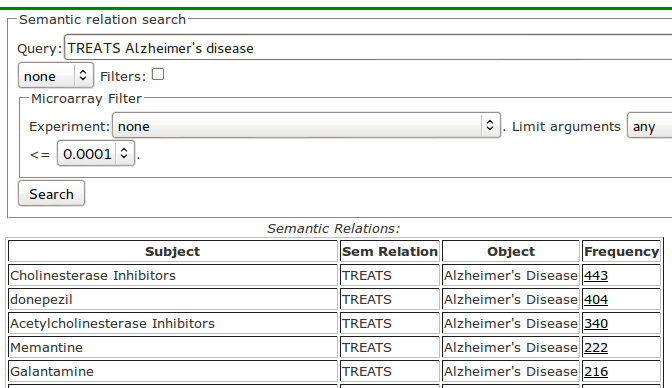
A more realistic question might ask "What are the treatments for Alzheimer's disease?" and can be specified in our tool in the simplest form as TREATS Alzheimer's disease as shown in Figure 2, in which some of the answers are: Donepezil-TREATS-Alzheimer's disease and Galantamine-TREATS-Alzheimer's disease.
The question "What does donepezil treat?" can be asked in our tool as donepezil TREATS and the answer will contain, in addition to Donepezil-TREATS-Alzheimer’s disease, other relations, including Donepezil-TREATS-Schizophrenia.
A very specific question such as "Whether donepezil has been used for Down syndrome?" could be asked as donepezil TREATS down syndrome in which all three components are specified; the returned relation Donepezil-TREATS-Down syndrome confirms that indeed it has been used for this disorder.
When specifying a question, the case does not matter: Donepezil TREATS Down syndrome is equivalent to donepezil treats down syndrome.
The semantic type of the subject and/or object can also be used in questions. For example, "Which pharmacologic substances cause which diseases or syndromes?" can be asked as phsu causes dsyn. Here phsu is the abbreviation of the semantic type "Pharmacological Substance" and dsyn is the abbreviation of the semantic type "Disease or Syndrome". When specifying a question in the current version of our tool, semantic types must be abbreviated; full names are not accepted.
A list of the semantic types and their corresponding abbreviations are shown in Table 1. The semantic relation names that can be used in questions are shown in Table 2.
| Abbreviation | Semantic Type |
|---|---|
| dsyn | Disease or Syndrome |
| aapp | Amino Acid, Peptide, or Protein |
| podg | Patient or Disabled Group |
| bpoc | Body Part, Organ, or Organ Component |
| gngm | Gene or Genome |
| topp | Therapeutic or Preventive Procedure |
| neop | Neoplastic Process |
| cell | Cell |
| mamm | Mammal |
| phsu | Pharmacologic Substance |
| orch | Organic Chemical |
| bacs | Biologically Active Substance |
| fndg | Finding |
| patf | Pathologic Function |
| popg | Population Group |
| aggp | Age Group |
| tisu | Tissue |
| celc | Cell Component |
| humn | Human |
| sosy | Sign or Symptom |
| diap | Diagnostic Procedure |
| orgf | Organism Function |
| inpo | Injury or Poisoning |
| celf | Cell Function |
| mobd | Mental or Behavioral Dysfunction |
| Predicate |
|---|
| PROCESS_OF |
| LOCATION_OF |
| PART_OF |
| TREATS |
| ISA |
| COEXISTS_WITH |
| AFFECTS |
| INTERACTS_WITH |
| USES |
| ASSOCIATED_WITH |
| CAUSES |
| ADMINISTERED_TO |
| STIMULATES |
| INHIBITS |
| AUGMENTS |
In the examples shown above we did not refer to the subject, relation and/or object explicitly, but rather implicitly. A query such as donepezil treats down syndrome searches all the words in all the fields of the relations. Most of the time, such a query will be satisfactory; however, it is possible to construct more precise queries by referring explicitly to particular search fields. Subject related search fields are:
sub_name meaning subject name
sub_semtype meaning subject semantic type abbreviation.
Object related search fields are:
obj_name meaning object name
obj_semtype meaning object semantic type abbreviation.
If we do not want to distinguish between the subject and the object, we can use: arg_name meaning the name of the subject or the object, and arg_semtype meaning the semantic type abbreviation of the subject or the object. And finally, there is one semantic relation related field - relation meaning the name of the relation. The query above with explicit search fields would look like sub_name:donepezil relation:treats obj_name:down syndrome.
Another implicit aspect of the queries shown so far is the logical connection or operator between the question terms. If there is no explicit logical operator present then AND is assumed. For example, the last query above really means and is equivalent to sub_name:donepezil AND relation:treats AND obj_name:down syndrome. When constructing questions, it is possible to use the standard Boolean operators AND, OR and NOT and group the search terms with parenthesis.
In contrast to the search terms themselves, the logical operators must be capitalized to be properly understood by the tool. The question "What are the genes or proteins known to be etiologically related to Alzheimer?" can be specified with explicit Boolean operators as sub_semtype:(aapp OR gngm) AND relation:(CAUSES OR PREDISPOSES OR ASSOCIATED_WITH) AND obj_name:Alzheimer where aapp stands for "Amino acid, peptide or protein" and gngm for "Gene or genome". An example of the NOT operator might be the question "What has been used to treat Alzheimer that is not a pharmacological substance?" which could be minimally specified as NOT phsu treats Alzheimer and in full form as NOT sub_semtype:phsu AND relation:treats AND obj_name:Alzheimer. As practical advice, we recommend that users of our QA tool first try specifying questions without explicit field reference and Boolean operators.
Automatic argument expansion is another useful feature of our QA tool. If requested, it expands the question arguments with semantically narrower concepts (hyponyms). For example, if we issue the query arg_name:antipsychotic treats we will get only relations where antipsychotic agents appears. However, if we use argument expansion by selecting from the Expand set of options before the query is submitted, the semantic relation ISA (meaning "is a") is used behind the scenes to search for narrower concepts, and the original query is expanded with them. The results will then also contain particular antipsychotic agents, such as clozapine, olanzapine, risperidone, haloperidol and so on. As another example, we can deal with a whole class of disorders in a question such as "What are the most common treatments for neurodegenerative disorders?" This question can be answered by using expansion in the query treats arg_name:neurodegenerative. Here, "neurodegenerative" is expanded with the particular neurodegenerative disorders, such as Alzheimer’s disease, Parkinson disease and so on.A similar question might be "What are the most common treatments for various neoplasms?" Here again we require expansion and use the query treats arg_name:neoplasms.
Currently, there are some limitations in the argument expansion facility:
arg_name, sub_name or obj_name).The last limitation means that when using expansion, the single word entered (e.g. "antipsychotic") is used to search for all the concepts containing that word (e.g. "antipsychotic agents", "atypical antipsychotic", "Antipsychotic Medications", ...), and, finally, all the concepts found are expanded. Therefore, although a single word is entered, it is possible to expand on multiple word concepts. These limitations are due to technical issues faced when parsing and modifying the original query, and we plan to remove them in the future.
When the user question is not specific enough at the beginning or when a more exploratory approach is taken, faceting is another promising avenue to explore. In our tool, faceting is turned on with the Filter option and is used for two purposes:
Faceting results are shown in the left column of the user interface (Figure 3).
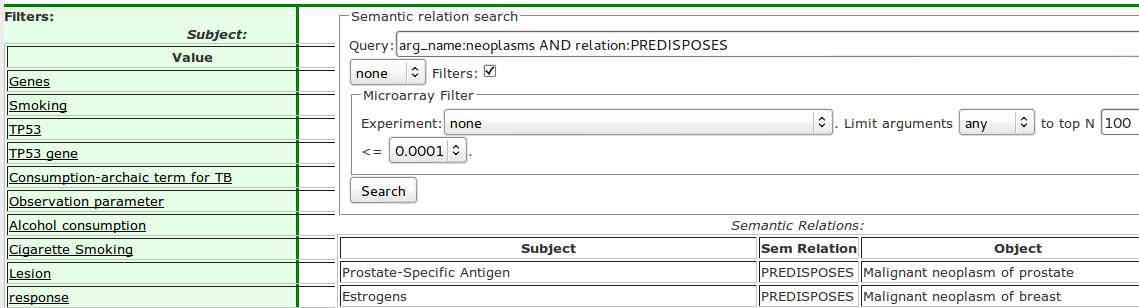
In our faceting approach top-N means, in case of the subjects, the top-N subjects by the number of relations in which they appear. In other words, a concept that appears as a subject most often in the semantic relations that are the answers to the original query will be shown at the top of the subject facet. The same method applies to the relation and object facets. For example, if the user wants to do some exploratory research on neoplasms and enters the query arg_name:neoplasms and also uses argument expansion the most common neoplasms are automatically included in the question. This is a very general question that results in several hundred thousand semantic relations. Now the user can browse the facets in the left column and investigate the subject, relations and objects appearing in highest number of relations. In the relation facet, the PREDISPOSES relation is selected in the relation facet, because that is the aspect the user wants to investigate further. The original query is automatically refined with the selected relation to become arg_name:neoplasms AND relation:PREDISPOSES (Figure 3). Now the results of the query show which concepts are known to predispose which particular neoplasms. The facets in the left column can be interpreted as:
The concepts in the subject facet are those that predispose the largest number of neoplasms.
The concepts in the object facet are the neoplasms with the largest number of known factors that predispose them.
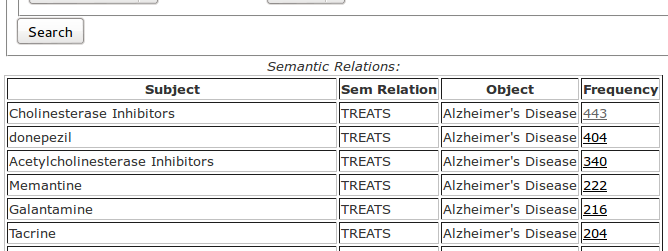
In the question processing phase the question entered by the user is interpreted depending on user-selected options; then it is executed. Answers are presented in a top-down fashion, semantic relations first, then, on demand, semantic relation instances, and finally, MEDLINE citations. In Figure 4 is the list of semantic relations, which are presented first. In addition to the subject, relation and object fields, the table also contains a Frequency field which is the number of instances of each relation in the table. The relations in the answer list are sorted by frequency of descending relation instance. In other words, the most frequent relation is at the top of the list.
The frequency field is a hyperlink and if followed, a new browser window shows the relation instances and a list of sentences from which each relation was extracted. In the sentences, whenever possible, the subject, relation and object are highlighted in different colors to make it easier to identify the relation and its context. Figure 5 shows the list of highlighted sentences for the semantic relation Donepezil-TREATS-Alzheimer’s disease. The highlighted sentences are listed in ascending order of argument-predicate distance, which is measured as the number of noun phrases between the arguments (subject and object) and the word indicating the semantic relation (the predicate).
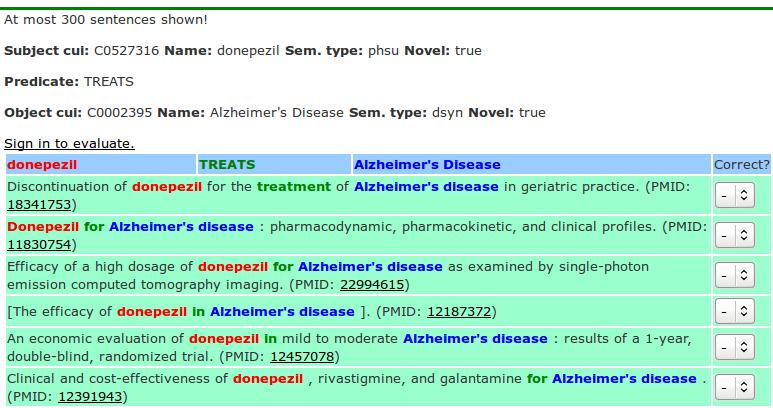
It is important to notice that the highlighted terms are not always the same as the official names used for the subject, relation or object. For example, in some sentences the abbreviation AD appears, but SemRep correctly recognizes this as Alzheimer's disease. Also, the words "in" and "for" are used several times in the text to indicate treatment, which is quite frequent in medical text. This is even more common when gene symbols are mentioned in the text. Many genes have more than one symbol to denote them. And often, different genes might have the same symbol. To make things even more difficult, some gene symbols can also have another, often more common, meaning. For example, CT and MR are gene symbols, but more often mean Computed Tomography and Magnetic Resonance, respectively. This problem is known as gene symbol ambiguity and SemRep attempts to address it as described in the Background section of the paper. At the end of each highlighted sentence, the PMID of the MEDLINE citation in which the sentence appears is shown as a link that, when followed, opens the MEDLINE citation so that the context of the sentence can be seen.